causalKNN Bootstrap Vignette
Walter Zhang
September 18, 2018
Source:vignettes/causalKNN-Bootstrap.Rmd
causalKNN-Bootstrap.RmdIn this vignette, we show how to utilize the bootstrap options in the
causalKNN package get a bagged estimate of our conditional
average treatment effect (CATE). This vignette extends the introductory
vignette.
Introduction
With the causal KNN, we can first bootstrap the procedure where we search for the optimal K and then the treatment effect estimation procedure. We will use the following simulated data set for our procedure.
set.seed(1234L)
n <- 5000 # Number of Observations in training set
p <- 21 # Number of Features
key <- 1:n # Individual indices
bootstrap_iterations <- 5L # Number of bootstrap iterations
n_test <- 501 # Number of Observations in test set
key_test <- 1:n_test
X <- cbind(matrix(rnorm(p/3*n), n, p/3),
matrix(5*runif(p/3*n), n, p/3),
matrix(rexp(p/3*n), n, p/3))
X_test <- matrix(0, n_test, p)
X_test[,1] <- rnorm(n_test)
X_test[,p/3+1] <- 5*runif(n_test)
TE <- pmin(abs(X[,1] * X[,p/3+1] + 1), 4)
W <- rbinom(n, 1, 0.5)
Y <- TE * W + 5 * X[,2] + X[,3] * pmax(X[,5], 0) + 5 * sin(X[,4]) + X[,8] + rnorm(n)
k <- 160L # Number of NN to search
distance_metric <- "euclidean" # Distance metric
keep_dist <- FALSE # Drop the distance matrices
threads <- 1L # Number of threads to use
knn_index_list <- knn_index_mat(X, W, k,
distance_metric = distance_metric,
keep_dist = keep_dist,
threads = threads)Searching for optimal K
N_step <- 20 # How many steps to take
K_step <- 5 # Value between steps and starting valueWe first bootstrap the keys and then supply the bootstrapped keys to the function.
boot_search_list <- lapply(1:bootstrap_iterations, function(bootstrap_index)
{
boot_key <- sample(key, length(key), replace = TRUE)
knn_optimal_k(X, W, Y, key, N_step, K_step, knn_index_list,
bootstrap_keys = boot_key)
})We then average across the RMSE values to find the optimal K value across the bootstrap results.
rmse_avg_vec <- rowMeans(sapply(boot_search_list, function(x) x$rmse_vec))
rmse_avg_vec[which.min(rmse_avg_vec)]
#> KNN_100
#> 214.5634
plot(boot_search_list[[1]]$k_vec, rmse_avg_vec, type = "l", ylab = "RMSE", xlab = "K")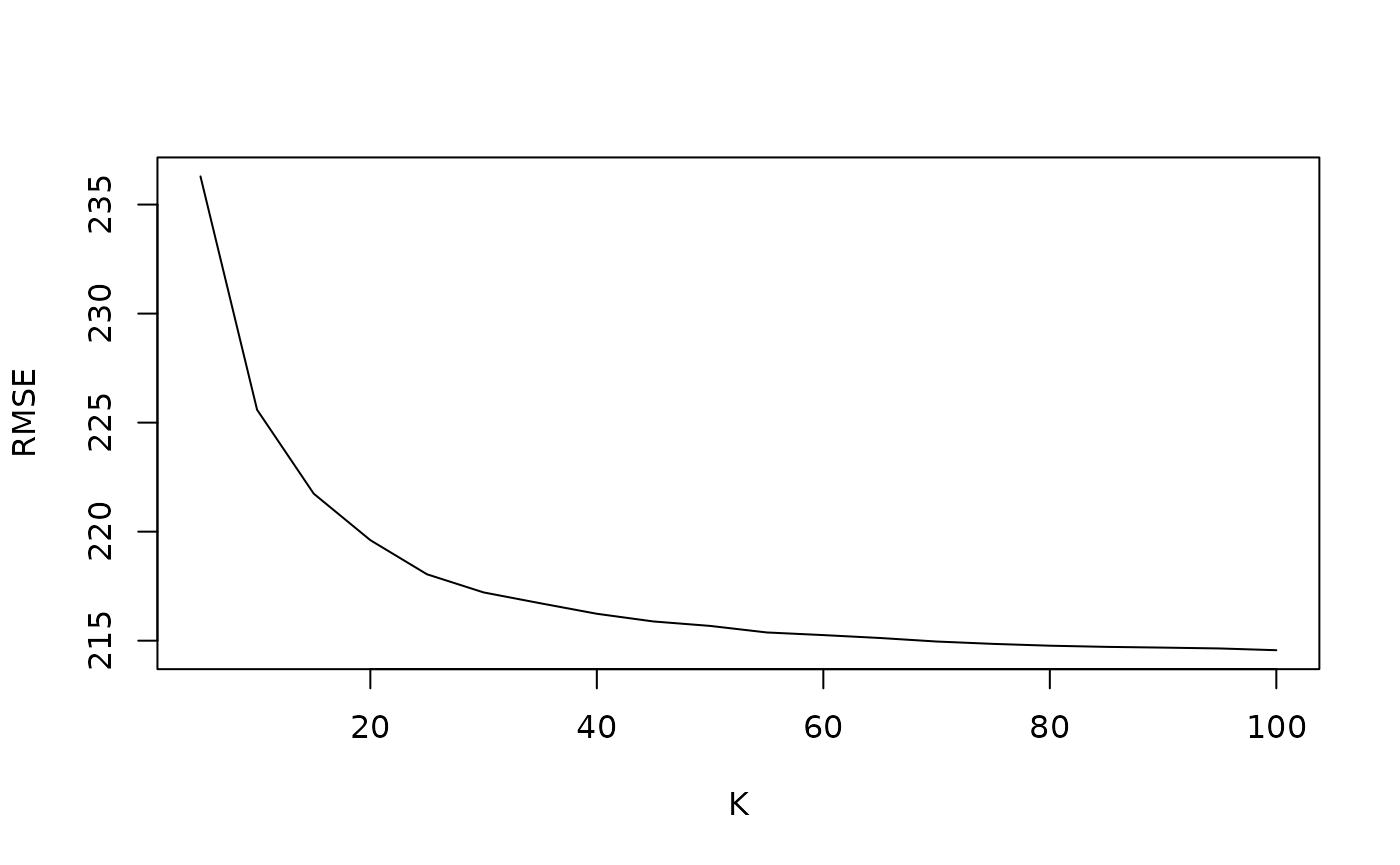
In this case, we see the bootstrapped optimal K search procedure gives us an optimal K value of \(100\). Note that this is the upper bound of our K grid. For production, we should try the procedure with a larger K grid to ensure we have reached the true optimal K value. For this vignette, we will use this K value of \(100\) for demonstrative purposes.
bagged_optimal_K <- 100LEstimating CATE
We can also use the bootstrap procedure to estimate the CATE in both the direct causal KNN and the TEP procedures.
Direct Causal KNN
We need to first compute the test set K nearest neighbor matrix.
knn_index_test_list <- knn_index_mat(X, W, k,
DF_test = X_test,
distance_metric = distance_metric,
keep_dist = keep_dist,
threads = threads)Then, we can average across bootstrap iterations our direct Causal KNN estimates.
boot_direct_list <- lapply(1:bootstrap_iterations, function(bootstrap_index)
{
boot_key <- sample(key, length(key), replace = TRUE)
causalknn_treatment_effect(W, Y, key, bagged_optimal_K,
knn_index_test_list,
key_test = key_test,
bootstrap_keys = boot_key)
})
direct_CATE_mat <- sapply(boot_direct_list, function(x) x$treatment_effect)
direct_CATE_DF <- data.frame(key = boot_direct_list[[1]]$key,
CATE = rowMeans(direct_CATE_mat))Treatment Effect Projection
We will our bootstrap our TEP projection estimates and then project using the Ridge for each bootstrap iteration. Then we average the Ridge’s predicted values for our bootstrap averaged causal KNN TEP estimates.
boot_TEP_list <- lapply(1:bootstrap_iterations, function(bootstrap_index)
{
boot_key <- sample(key, length(key), replace = TRUE)
causal_KNN_DF <- causalknn_treatment_effect(W, Y, key, bagged_optimal_K,
knn_index_list,
bootstrap_keys = boot_key)
TEP_results_list <- tep_enet(X, causal_KNN_DF$treatment_effect,
X_test, alpha = 0)
list(causal_KNN_DF = causal_KNN_DF, TEP_results_list = TEP_results_list)
})
TEP_CATE_mat <- sapply(boot_TEP_list, function(x) x$TEP_results_list$test_TE)
TEP_CATE_DF <- data.frame(key = key_test,
CATE = rowMeans(TEP_CATE_mat))